Python is a powerful programming language that is easy to learn and easy to work with, but it is not always the fastest to run—especially when you’re dealing with math or statistics. Third-party libraries like NumPy, which wrap C libraries, can improve the performance of some operations significantly, but sometimes you just need the raw speed and power of C directly in Python.
Cython was developed to make it easier to write C extensions for Python, and to allow existing Python code to be transformed into C. What’s more, Cython allows the optimized code to be shipped with a Python application without external dependencies.
In this tutorial we’ll walk through the steps needed to transform existing Python code into Cython, and to use it in a production application.
A Cython example
Let’s begin with a simple example taken from Cython’s documentation, a not-very-efficient implementation of an integral function:
def f(x):
return x**2-x
def integrate_f(a, b, N):
s = 0
dx = (b-a)/N
for i in range(N):
s += f(a+i*dx)
return s * dx
The code is easy to read and understand, but it runs slowly. This is because Python must constantly convert back and forth between its own object types and the machine’s raw numerical types.
Now consider the Cython version of the same code, with Cython’s additions underscored:
cdef f(double x):
return x**2-x
def integrate_f(double a, double b, int N):
cdef int i
cdef double s, x, dx
s = 0
dx = (b-a)/N
for i in range(N):
s += f(a+i*dx)
return s * dx
These additions allow us to explicitly declare variable types throughout the code, so that the Cython compiler can translate those “decorated” additions into C.
Cython syntax
The keywords used to decorate Cython code are not found in conventional Python syntax. They were developed specifically for Cython, so any code decorated with them won’t run as a conventional Python program.
These are the most common elements of Cython’s syntax:
Variable types
Some of the variable types used in Cython are echoes of Python’s own types, such as int, float, and long. Other Cython variable types are also found in C, like char or struct, as are declarations like unsigned long. And others are unique to Cython, like bint, a C-level representation of Python True/False values.
The cdef and cpdef function types
The cdef keyword indicates the use of a Cython or C type. It is also used to define functions much as you would in Python.
Functions written in Cython using Python’s def keyword are visible to other Python code, but incur a performance penalty. Functions that use the cdef keyword are only visible to other Cython or C code, but execute much faster. If you have functions that are only called internally from within a Cython module, use cdef.
A third keyword, cpdef, provides compatibility with both Python code and C code, in such a way that C code can access the declared function at full speed. This convenience comes at a cost, though: cpdef functions generate more code and have slightly more call overhead than cdef.
Other Cython keywords
Other keywords in Cython provide control over aspects of program flow and behavior that isn’t available in Python:
gilandnogil. These are context managers used to delineate sections of code that require (with gil:) or do not require (with nogil:) Python’s Global Interpreter Lock, or GIL. C code that makes no calls to the Python API can run faster in anogilblock, especially if it is performing a long-running operation.cimport. This directs Cython to import C data types, functions, variables, and extension types. Cython apps that use NumPy’s native C modules, for instance, usecimportto gain access to those functions.include. This places the source code of one Cython file inside another, in much the same way as in C. Note that Cython has a more sophisticated way to share declarations between Cython files other than justincludes.ctypedef. Used to refer to type definitions in external C header files.extern. Used withcdefto refer to C functions or variables found in other modules.public/api. Used to make declarations in Cython modules that will be visible to other C code.inline. Used to indicate a given function should be inlined, or have its code placed in the body of the calling function whenever it is used, for the sake of speed. For instance, theffunction in the above code example could be decorated withinlineto reduce its function call overhead, because it is only used in one place.
It is not necessary to know all of the Cython keywords in advance. Cython code tends to be written incrementally—first you write valid Python code, then you add Cython decoration to speed it up. Thus you can pick up Cython’s extended keyword syntax piecemeal, as you need it.
Compile Cython
Now that we have some idea of what a simple Cython program looks like and why it looks the way it does, let’s walk through the steps needed to compile Cython into a working binary.
To build a working Cython program, we will need three things:
- The Python interpreter. Use the most recent release version, if you can.
- The Cython package. You can add Cython to Python by way of the
pippackage manager:pip install cython - A C compiler.
Item #3 can be tricky if you’re using Microsoft Windows as your development platform. Unlike Linux, Windows doesn’t come with a C compiler as a standard component. To address this, grab a copy of Microsoft Visual Studio Community Edition, which includes Microsoft’s C compiler and costs nothing.
Cython programs use the .pyx file extension. In a new directory, create a file named num.pyx that contains the Cython code example shown above (the second code sample under “A Cython example”) and a file named main.py that contains the following code:
from num import integrate_f
print (integrate_f(1.0, 10.0, 2000))
This is a regular Python progam that will call the integrate_f function found in num.pyx. Python code “sees” Cython code as just another module, so you don’t need to do anything special other than import the compiled module and run its functions.
Finally, add a file named setup.py with the following code:
from distutils.core import setup
from distutils.extension import Extension
from Cython.Build import cythonize
ext_modules = [
Extension(
r’num’,
[r’num.pyx’]
),
]
setup(
name=’num’,
ext_modules=cythonize(ext_modules),
)setup.py is normally used by Python to install the module it’s associated with, and can also be used to direct Python to compile C extensions for that module. Here we’re using setup.py to compile Cython code.
If you’re on Linux, and you have a C compiler installed (typically the case), you can compile the .pyx file to C by running the command:
> python setup.py build_ext --inplace
If you’re using Microsoft Windows, you’ll want to add a batch file named compile.bat to automate the compilation process:
@SETLOCAL
set DISTUTILS_USE_SDK=1
call “C:\Program Files (x86)\Microsoft Visual Studio\2017\Community\VC\Auxiliary\Build\vcvarsall.bat” amd64
python setup.py build_ext --inplace --compiler=msvc
Note that the exact path to vcvarsall.bat in line 3 will vary depending on the version of Visual Studio you have installed. This example assumes you’re using Visual Studio 2017 Community as per the earlier direction.
If the compilation is successful, you should see new files appear in the directory: num.c (the C file generated by Cython) and a file with either a .o extension (on Linux) or a .pyd extension (on Windows). That’s the binary that the C file has been compiled into. You may also see a \build subdirectory, which contains the artifacts from the build process.
Run python main.py, and you should see something like the following returned as a response:
283.297530375
That’s the output from the compiled integral function, as invoked by our pure Python code. Try playing with the parameters passed to the function in main.py to see how the output changes.
Note that whenever you make changes to the .pyx file, you will need to recompile it. Of course, any changes you make to conventional Python code will be affected immediately.
The resulting compiled file has no dependencies except the version of Python it was compiled for, and so can be bundled into a binary wheel. Note that if you refer to other libraries in your code, like NumPy (see below), you will need to provide those as part of the application’s requirements.
How to use Cython
Now that you know how to “Cythonize” a piece of code, the next step is to determine how your Python application can benefit from Cython. Where exactly should you apply it?
For best results, use Cython to optimize these kinds of Python functions:
- Functions that run in tight loops, or require long amounts of processing time in a single “hot spot” of code.
- Functions that perform numerical manipulations.
- Functions that work with objects that can be represented in pure C, such as basic numerical types, arrays, or structures, rather than Python object types like lists, dictionaries, or tuples.
Python has traditionally been less efficient at loops and numerical manipulations than other, non-interpreted languages. Issue #3 is a byproduct of issue #2: the more you can decorate your code to indicate it should use base numerical types that can be turned into C, the faster it’ll be able to do number-crunching.
Using Python object types in Cython isn’t itself a problem. Cython functions that use Python objects will still compile, and Python objects may be preferable when performance isn’t the top consideration. But any code that makes use of Python objects will be limited by the performance of the Python runtime, as Cython will generate code to directly address Python’s APIs and ABIs.
Another worthy target of Cython optimization is Python code that interacts directly with a C library. You can skip the Python “wrapper” code and interface with the libraries directly.
However, Cython does not automatically generate the proper call interfaces for those libraries. You will need to have Cython refer to the function signatures in the library’s header files, by way of a cdef extern from declaration. Note that if you don’t have the header files, Cython is forgiving enough to let you declare external function signatures that approximate the original headers. But use the originals whenever possible to be safe.
One external C library that Cython can use right out of the box is NumPy. To take advantage of Cython’s fast access to NumPy arrays, use cimport numpy (optionally with as np to keep its namespace distinct), and then use cdef statements to declare NumPy variables, such as cdef np.array or np.ndarray.
Cython profiling
The first step to improving an application’s performance is to profile it—to generate a detailed report of where the time is being spent during execution. Python provides built-in mechanisms for generating code profiles. Cython not only hooks into those mechanisms but has profiling tools of its own.
Python’s own profiler, cProfile, generates reports that show which functions take up the most amount of time in a given Python program. By default, Cython code doesn’t show up in those reports, but you can enable profiling on Cython code by inserting a compiler directive at the top of the .pyx file with functions you want to include in the profiling:
# cython: profile=True
You can also enable line-by-line tracing on the C code generated by Cython, but this imposes a lot of overhead, and so is turned off by default.
Note that profiling imposes a performance hit, so be sure to toggle profiling off for code that is being shipped into production.
Cython can also generate code reports that indicate how much of a given .pyx file is being converted to C, and how much of it remains Python code. To see this in action, edit the setup.py file in our example and add the following two lines at the top:
import Cython.Compiler.Options
Cython.Compiler.Options.annotate = True
Delete the .c files generated in the project and re-run the setup.py script to recompile everything. When you’re done, you should see an HTML file in the same directory that shares the name of your .pyx file—in this case, num.html. Open the HTML file and you’ll see the parts of your code that are still dependent on Python highlighted in yellow. You can click on the yellow areas to see the underlying C code generated by Cython.
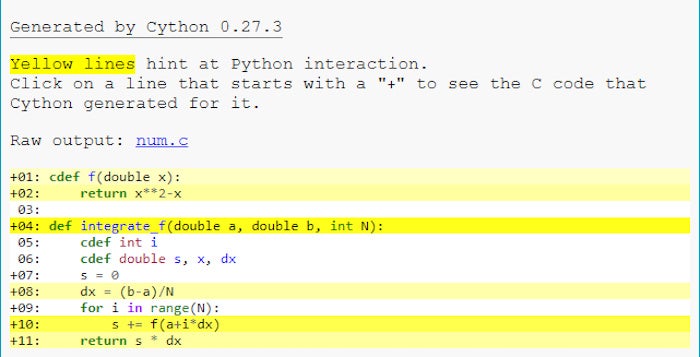 IDG
IDG
A Cython code report. The yellow highlights indicate parts of the code that still depend on the Python runtime.
In this case, the cdef f function is still highlighted in yellow, despite being a cdef function and having its variable explicitly typed. Why? Because the return value of the function isn’t typed. Cython assumes that the function will return a Python object, not a double, so has generated Python API code to handle that.
You can fix this by editing the cdef f declaration to read:
cdef double f(double x):
Save and recompile the file, and reload the report. You should now see the cdef f function is no longer highlighted in yellow; it’s pure C.
 IDG
IDG
The revised function, now pure C, generates no highlights.
Note that if you have profiling enabled as described above, even “pure” C functions will show some highlighting, because they have been decorated with trace code that calls back into Python’s internals.






